A landmark study in the field of food allergies made in Luxembourg
26 July 2022
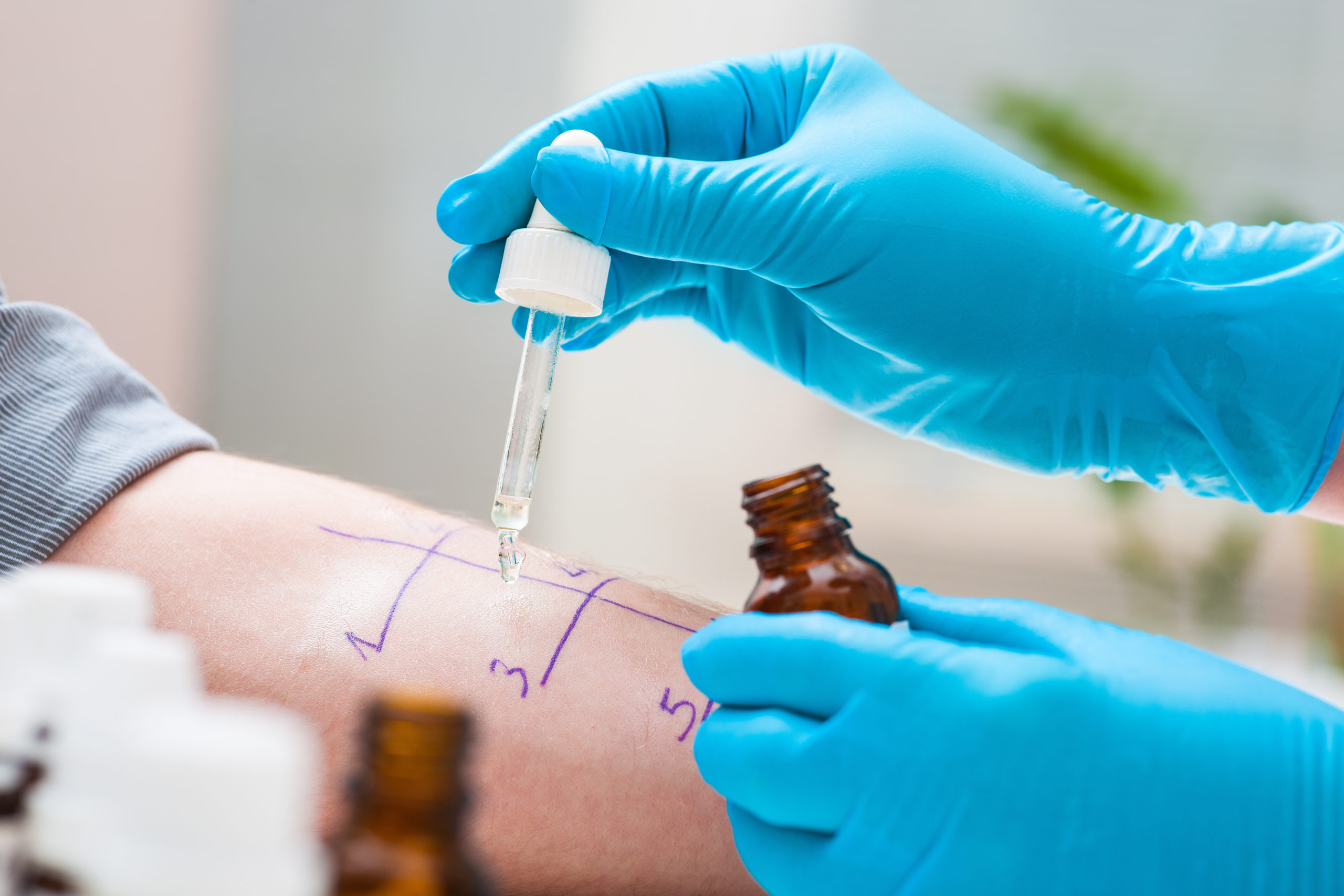
Mapping Real-Time Allergic Responses.
While allergies affect up to 30% of people today, largely in children and young people, an all Luxembourg study led by the Luxembourg Institute of Health (LIH) has begun to unravel the real-time immune response of subjects suffering from peanut allergies.
Researchers at the Luxembourg Institute of Health (LIH) begin to reveal the body’s step-by-step immune response to peanut allergies
A first of its kind investigation
This all Luxembourg study has begun to unravel the real-time immune response of subjects suffering from peanut allergies. The first of its kind investigation was able to identify biological markers in children that could help clinicians to assess how and under what circumstances a child might react, with future implications for better treatments and prevention strategies.
Public awareness of health and disease has never been more pronounced than in recent years, with infectious diseases, cancer and mental health all being high on the media and government agendas. It is interesting therefore that the most common chronic disease today doesn’t fall into any of these categories, and has been quietly growing without any particular fanfare.
A pressing need for new therapeutic and diagnostic solutions
Allergies affect up to 30% of people today, largely in children and young people, with the rising numbers showing little signs of slowing down. Treatment options are also fairly restricted, meaning that there is an increasing burden on health systems and a pressing need for new therapeutic and diagnostic solutions. Dr Annette Kuehn and colleagues at the LIH’s Department of Infection and Immunity (DII) have been seeking to address this need by monitoring immune reactions before and during the onset of allergic symptoms upon peanut ingestion.

“Food allergies have been skyrocketing over the last two decades and there is no end in sight for that prevalence increase. Initially, this was mostly a pediatric disease (ca. 5% in pre-schoolers) but today, more and more adults are affected as well. Therapeutic options are limited so there is a great need for action in this domain.”
Dr Annette Kuehn
One of the major problems with food allergies is that they manifest in an extremely personalised fashion. Some patients experience symptoms in the gastrointestinal tract, some on the skin, others severe acute asthma or swelling in the throat, with varying degrees of severity based on different thresholds of the allergen. The food allergy severity on an individual basis is difficult to predict. As a result, this can make the job of clinicians trying to assess an allergic response very difficult.
Identify markers to predict peanut exposure’s response
In the new study led by Dr Kuehn and Prof. Markus Ollert, in close collaboration with the medical partner Dr Morel-Codreanu from the National Allergology-Immunology Unit at the Centre Hospitalier de Luxembourg (CHL), the aim was to search for different biological markers in the blood of children with suspected peanut allergies throughout a controlled exposure to peanuts. By doing this, the goal of the researchers was to identify markers that would enable them to predict how an individual might respond to peanut exposure. These predictions could include not only whether or not a child would react to peanuts, but with which symptoms and based on how much peanut. This could therefore act as a powerful diagnostic tool for clinicians, removing the need for laborious and costly approaches.
This was the first study of its kind to investigate this time dependent aspect of the immune response in human subjects, making it a landmark in the field. Twenty-six children with an average age of seven and a half years were assessed, as well as seven peanut-tolerant adults included as controls. Samples were taken from the participants both before, during and 1 hour after the onset of any symptoms after controlled doses of peanut had been administered under medical supervision at CHL.
Useful results in the the diagnosis and monitoring of allergic responses
From their results, the team was able to make some extremely encouraging conclusions. Firstly, that from the participants’ immune signatures, it was possible to discriminate between those with or without an allergic response. In the cases of an allergic response, it was generally seen that specific blood immune cells tended to be fewer in number, but with an increased expression of ‘homing markers’; indications that these cells are better equipped to enter affected tissues resulting in symptoms. Those patients without an allergic response despite having received a sufficiently high allergen dose still showed signs of ongoing inflammation, but in a much more controlled manner, with cells less able to penetrate affected tissues.
A second success was the ability to identify groups of children that tolerated higher or lower doses of peanuts before developing an allergic response, based on variations in the abundance of certain white blood cells and other key immune cells. The results were even able to determine whether the symptoms experienced were primarily related to the skin or gastrointestinal tract. As such, this meant that this kind of analysis could prove extremely useful in the diagnosis and monitoring of allergic responses.

“This is the first in-human study to directly analyse the in-vivo immune response during allergic reactions to food. Not only could blood biomarkers help to reduce diagnostic food challenges, but with the knowledge we gain by analysing our immune response in-vivo, it may ultimately enable us to better treat and prevent food allergies in the future.”
Rebecca Czolk, PhD student and study co-author
The study was recently published in the highly acclaimed academic journal ‘Allergy’, under the full title “High-dimensional immune profiles correlate with phenotypes of peanut allergy during food-allergic reactions” (DOI: 10.1111/all.15408).
Funding and collaborations
This study was supported by the Luxembourg National Research Fund on PRIDE program grants PRIDE/11012546/NEXTIMMUNE (JK, GM, CH, MO, AK) and PRIDE17/11823097/MICROH (RC, AK); supported by the Personalized Medicine Consortium grant APSIS, PMC/2017/02 (AK) and by intramural funds of the Ministry of Higher Education and Research (MESR), Luxembourg (AC, CH, MO, AK).










